
MONMOUTH, IL (January 24, 2019) — Monmouth College students continue to contribute to a "tremendous resource to the scientific community" by helping unlock mysteries about a virus.
Phage are viruses that infect bacteria, and they "are everywhere that bacteria are found," said Monmouth biology professor Eric Engstrom, who is working with Monmouth students on learning more about phage, along with department colleagues Tim Tibbetts and James Godde.
During the fall semester, students isolated phage, finding it on campus near Fulton Hall and at the College's LeSuer Nature Preserve. Once the phage was isolated from the mixture of soil, bacteria and other phages, phage was grown to a sufficient titer, which is the number of phage in a potential unit.
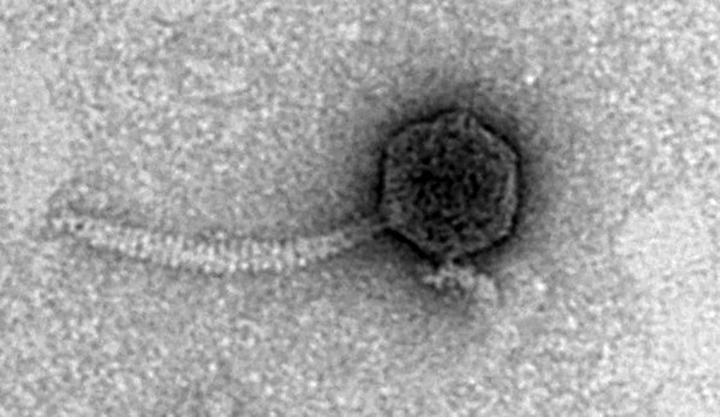
That growth allowed for an exciting next step, as four samples have been genetically sequenced. The students also have "some fantastic images" of the samples through the University of Iowa's imaging facility.
Monmouth samples in national database
"They're in there — they're archived," said Engstrom of the samples in the Actinobacteriophage Database at PhagesDB.org, an interactive site that collects and shares information related to the discovery, characterization and genomics of phages that infect bacterial hosts. "It's a very high-quality database that continues to improve with more annotation. It's a tremendous resource to the scientific community."
Monmouth students aren't the only undergraduates sending samples to the database. Administered by the Howard Hughes Medical Institute, the Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES), is a nationwide discovery-based undergraduate research initiative. Other schools participating range from community colleges to large research institutions such as Johns Hopkins University.
"They're fascinating things, but we don't know how to think about phage," said Engstrom. "We're trying to broaden our understanding. Most of its genes, we have no idea what they do. This opens up a lot of questions, such as are they phage-specific? We just don't know until we have lots of samples."
Research was conducted in the 1950s on using phage to treat infection, but that information was pushed to the back burner because of the rise of antibiotics. Phage has become relevant again because some diseases, such as tuberculosis, are resisting antibiotics.
"We're becoming interested in this again," Engstrom said last fall. "We let it rust. Can we clean it up and put it back to use?"
This spring's lab work
Students who hunted phage during an introductory laboratory course in the fall are now working on "annotating" their samples, a process Engstrom said could take from six weeks to the entire spring semester.
Although a computer, known as the Phamerator, can do gene annotation, Engstrom said humans have an advantage.
"Computers can do a crude job, but it's not good enough," he said. "The process really requires human eyes, a human brain. ... The better we get at this, we can fix the errors. We're a part of that process here in our lab."
Within the process, Engstrom said three questions are posed: "Is there a gene here? Where does the gene start? Where it ends is easy to discern, but the start can be very ambiguous. And what does the gene do - are there assignable functions?"
He said those questions open up fascinating science.
"For many of the genes, the functions are unknown. It gives us new things to figure out."
The students are working in groups of three on the annotation process. The first few labs of the semester have served as a review, as well as an introduction to Phamerator and other software.
Looking ahead
When the annotation process is complete, Monmouth students will be listed as "authors for each GenBank entry," said Engstrom. Produced and maintained by the National Center for Biotechnology Information, the GenBank sequence database is an open-access, annotated collection of all publicly available nucleotide sequences and their protein translations.
Two Monmouth students will be selected to attend the 12th annual meeting of SEA-PHAGES in June at the Howard Hughes Medical Institute's Janelia Research Campus near Ashburn, Virginia.
Engstrom expects about 20 members of next fall's incoming freshman class to participate in the next phage-hunting lab. He said it's possible that in years to come, the phage lab will be the default biology lab for all incoming students, "but we're not there yet."
"The very first thing they encounter is real, functional science," Engstrom said of freshman students working in the phage lab. "This is an authentic research lab. It's different from the typical lab experience. We're getting new knowledge and learning about the world."










